The -not so new- demanding situations when speaking about PRRS virus
It’s not information that porcine reproductive and breathing syndrome virus (PRRSV) has necessary options that give a contribution to consistent illness re-emergence in swine herds which ends up in really extensive animal well being problems and financial losses. This sort of options is its speedy capability to mutate and doubtlessly grow to be extra serious and/and even infect animals in the past “immune” to another PRRSV variant. Because of this, with the intention to perceive a PRRSV “kind” found in a farm – whether or not that could be a virus from a brand new outbreak or a “resident” virus- it turned into quite common to investigate the PRRSV genome.
This process is normally achieved by means of sequencing crucial a part of PRRSV known as the ORF5 gene; to know the place the virus suits evolutionarily and the way it compares to earlier virus that had been detected within the web page or locally/ area.
PRRSV sequencing data has been broadly used within the box to:
- perceive which virus is inflicting issues
- for steerage on outbreak supply
- for differentiation between vaccine traces and “dwelling”/ newly presented traces
- for vaccine possible choices, and so on.
Not too long ago, the “lineage” classification has been increasingly more used to categorise viruses. This system used to be evolved round 2010 (Shi et al., 2010), when PRRSV had been divided into 9 distinct lineages; and additional delicate when PRRSV used to be additional subdivided into sub lineages.
The elephant within the room: is sequencing one pattern just right sufficient?
PRRS sequencing is normally no longer affordable, due to this fact it’s extra usually achieved underneath “particular scenarios” and no longer mechanically (e.g. right through new outbreaks, after virus inoculation occasions, and so on.). As well as, right through those instances, just one pattern or a pool of samples (out of many usually taken on-farm) is used for sequencing within the overwhelming majority of diagnostic instances.
The implication is that, in lots of instances, veterinarians, manufacturers, and different animal well being workforce usually use a unmarried series data to make necessary conclusions relating to virus supply (particularly for brand spanking new outbreaks), information outbreak investigations, and to tell long run interventions.
In our learn about, we needed to verify that, if we sequenced a couple of samples taken inside of the similar day, all samples would yield the similar series, or sequences that will be “shut sufficient” to be regarded as a minimum of the similar virus lineage. We additionally sought after to try whole-genome sequencing (WGS) together with ORF5 sequencing to peer whether or not we might accumulate additional information. WGS can have a bonus because it offers data at the total virus, whilst ORF5 has been traditionally used however handiest tells us a work of the tale. Then again, since it’s nonetheless a rather contemporary building and will also be pricey (in america it prices roughly 3 times greater than the ORF5 sequencing), we’re nonetheless finding out interpret it and our “database” for comparability is way smaller in comparison to ORF5.
We suspected that, given the speedy mutation charge of PRRS and the huge measurement of contemporary swine amenities, shall we in finding a couple of lineages of PRRS inside of a sampling match, which might be regarding. A graphical abstract of our learn about rationale is in Fig. 1.
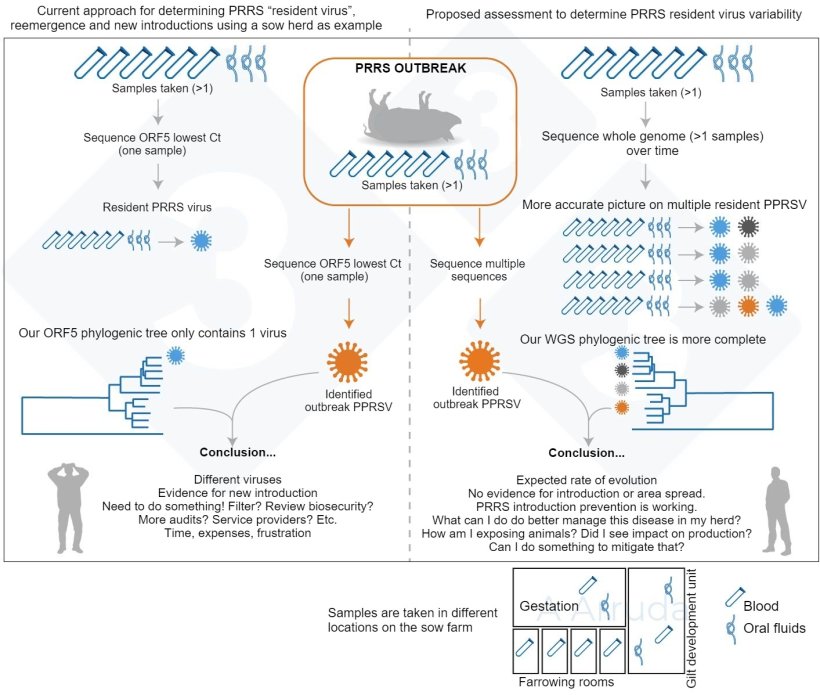
Finding out what number of PRRS traces are we able to in finding on our farms
We enrolled 5 farms in the learn about, 3 breeding herds and a pair of grow-finish; and sampled per month for about twelve months the use of several types of samples (tonsil scrapings, oral fluids, processing fluids). We all the time sampled the similar places within the barn that had been spatially unfold out all through the amenities to take a look at and get probably the most illustration conceivable of the entire web page. We had as much as 16 samples consistent with web page each month, and they all had been examined the use of quantitative PCR; and following this, the certain samples had been ORF5 sequenced.
What did we discover out?
Our maximum necessary discovering used to be that underneath box prerequisites, we had been ready to stumble on as much as 3 other PRRSV lineages right through a unmarried sampling match for breeding herds, and as much as two for rising pig herds. That is proven on Fig. 2.
Determine 2. PRRSV sub-lineages encountered in learn about farms (1-5) all through the length of our sampling occasions (in months) for the other pattern sorts.
| Sampling match | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Farm | Pattern kind | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 1 | Processing fluid | L1H | L1H | ||||||||||
| Tonsil scraping | |||||||||||||
| 2 | Processing fluid | L1H | L1H | L1A | L1H | L1H | |||||||
| L1H | |||||||||||||
| L8† | |||||||||||||
| Tonsil scraping | L1H | ||||||||||||
| 3 | Processing fluid | L1H | L1H | L1H | L1H | ||||||||
| Tonsil scraping | |||||||||||||
| 4 | Oral fluid | L5† | L1A | ||||||||||
| L5† | |||||||||||||
| Tonsil scraping | L1A | L5† | L5† | L1A | |||||||||
| L5† | |||||||||||||
| 5 | Oral fluid | L1A | L1A | L5† | |||||||||
| Tonsil scraping | L1A | L1A | |||||||||||
We additionally famous that, on the subject of our learn about, oral fluid samples had been very tricky to series, whilst processing fluids had been the very best. Taking into consideration each are “composite” samples, that might doubtlessly be defined by means of total upper Ct values, indicating much less viral quantities for oral fluids in comparison to processing fluids. We attempted to try WGS however in any case, we had been not able to procure complete whole-genome sequences from any of our samples. We suspect that may be because of the sorts of samples we applied, however additional analysis is wanted in this subject.
Those findings display how necessary it’s to imagine sequencing a number of samples when making necessary choices, even supposing we perceive this may occasionally include upper prices.
Acknowledgments
The Nationwide Red meat Board supplied investment for this mission. We wish to thank veterinarians and manufacturers who had been instrumental for pattern assortment.
